Difference between revisions of "Microbial research"
(→References) |
Dronkers J (talk | contribs) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 7: | Line 7: | ||
billion tonnes of carbon. In a drop (one millilitre) of seawater, one can find 10 million | billion tonnes of carbon. In a drop (one millilitre) of seawater, one can find 10 million | ||
viruses, one million bacteria and about 1,000 small protozoans and algae (called “protists”). | viruses, one million bacteria and about 1,000 small protozoans and algae (called “protists”). | ||
| − | In addition to their high abundance, microbes play a crucial role in most biogeochemical processes occurring in the marine environment: they account for almost half of global primary production and form a major part of ecosystem respiration and [[Nutrient | + | In addition to their high abundance, microbes play a crucial role in most biogeochemical processes occurring in the marine environment: they account for almost half of global primary production and form a major part of ecosystem respiration and [[Nutrient conversion in the marine environment|nutrient recycling]]<ref name="ma">[https://www.researchgate.net/publication/306030378_Marine_Biodiversity_and_Ecosystem_Functioning Heip, C., Hummel, H., van Avesaath, P., Appeltans, W., Arvanitidis, C., Aspden, R., Austen, M., Boero, F., Bouma, TJ., Boxshall, G., Buchholz, F., Crowe, T., Delaney, A., Deprez, T., Emblow, C., Feral, JP., Gasol, JM., Gooday, A., Harder, J., Ianora, A., Kraberg, A., Mackenzie, B., Ojaveer, H., Paterson, D., Rumohr, H., Schiedek, D., Sokolowski, A., Somerfield, P., Sousa Pinto, I., Vincx, M., Węsławski, JM., Nash, R. (2009). Marine Biodiversity and Ecosystem Functioning. Printbase, Dublin, Ireland ISSN 2009-2539]</ref>. |
<P> | <P> | ||
<BR> | <BR> | ||
| Line 20: | Line 20: | ||
differences in that gene enables organisms to be classified into different [[taxon|taxa]] in a way that reflects their evolutionary history. | differences in that gene enables organisms to be classified into different [[taxon|taxa]] in a way that reflects their evolutionary history. | ||
| − | Initial reports of microbial richness in aquatic environments suggested that there were less than 200 different microorganisms in a typical sea water sample. But recent advances in molecular technologies have shown that the diversity is far much greater. Today it is thought that a single seawater sample may contain up to 10.000 different types of microorganisms | + | Initial reports of microbial richness in aquatic environments suggested that there were less than 200 different microorganisms in a typical sea water sample. But recent advances in molecular technologies have shown that the diversity is far much greater. Today it is thought that a single seawater sample may contain up to 10.000 different types of microorganisms<ref name="ma"/>. |
<P> | <P> | ||
[[Image:Microbes.jpg|thumb|center|700px| <div style="text-align: center;"> | [[Image:Microbes.jpg|thumb|center|700px| <div style="text-align: center;"> | ||
| − | A conceptual model of how microbial diversity in the ocean is expressed, i.e. a few organisms are relatively abundant and participate in [[ecosystem functioning]] while the majority of microbes await more optimal growing conditions, as shown above in the large ‘tail of biodiversity’. Adaptation | + | A conceptual model of how microbial diversity in the ocean is expressed, i.e. a few organisms are relatively abundant and participate in [[Biodiversity, ecosystem functioning and ecosystem function|ecosystem functioning]] while the majority of microbes await more optimal growing conditions, as shown above in the large ‘tail of biodiversity’. Adaptation of a figure by Pedrós-Alió (2006).</div>]] |
<P> | <P> | ||
<BR> | <BR> | ||
| Line 31: | Line 31: | ||
== Specific discoveries== | == Specific discoveries== | ||
| − | To understand plankton distribution and changes, | + | To understand plankton distribution and changes, one should know how diverse micro-plankton species are. Some fundamental questions are: ‘Can a bacterium species evolve locally despite its limited dispersal?’ and conversely, ‘Is there a bacterium species which inhabits all European waters?’ |
| − | ''Rhodopirellula'', an abundant red bacterium that lives attached to marine sediment grains was selected to investigate these questions. 70 different strains, representing several different species of the ''Rhodopirellula'' genus were isolated. ''Rhodopirellula baltica'' was restricted to the [[Baltic Sea]], the Skagerrak and the Eastern [[North Sea]]. A second ''Rhodopirellula'' species of was present in Iceland and Scotland, representing the North- | + | ''Rhodopirellula'', an abundant red bacterium that lives attached to marine sediment grains was selected to investigate these questions. 70 different strains, representing several different species of the ''Rhodopirellula'' genus were isolated. ''Rhodopirellula baltica'' was restricted to the [[Baltic Sea]], the Skagerrak and the Eastern [[North Sea]]. A second ''Rhodopirellula'' species of was present in Iceland and Scotland, representing the North-Atlantic habitat. Another ''Rhodopirellula'' species was obtained from the Adriatic Sea, but the majority of the isolates belonged to a forth ''Rhodopirellula'' species which was present in the English Channel, on the French Atlantic coast and in the [[Mediterranean Sea|Mediterranean]]. This showed that different [[species]] of the ''Rhodopirellula'' genus evolved within the European seas. |
| Line 39: | Line 39: | ||
The trick is to find out “who is doing what” and “who is the most relevant” among those that perform a given biogeochemical function and what effect global change will have on that particular species. | The trick is to find out “who is doing what” and “who is the most relevant” among those that perform a given biogeochemical function and what effect global change will have on that particular species. | ||
<P> | <P> | ||
| − | Members of the MarBEF project | + | Members of the MarBEF project have researched the identity and ecological function of the [[keystone species|key]] microbial organisms in different areas<ref name="ma"/>: the central Baltic Sea, the North West Mediterranean Sea and the deep North Atlantic. |
| − | + | ||
| − | + | ||
| − | |||
==Microbial research in the central Baltic Sea== | ==Microbial research in the central Baltic Sea== | ||
| − | |||
===Anoxic zones=== | ===Anoxic zones=== | ||
| + | [[Anoxic]] bottom waters are characteristic features of marginal and enclosed seas and many coastal environments ([[Black Sea]], [[Baltic Sea]], [[fjord|fjords]], etc) and are increasing in extent worldwide. | ||
| − | + | In oxic-anoxic transition zones, elements are transformed from one state into another. These transformations have an impact the biogeochemical cycles and are important on an ecosystem scale. Furthermore, these environments can give insight in how ancient oceans, which were mostly anoxic throughout much of the Earth’s history, worked. These transition zones are also ideal sites to study the link between microbial community structure and biogeochemical transformations (and thus between [[Marine Biodiversity|biodiversity]] and [[Biodiversity, ecosystem functioning and ecosystem function|ecosystem function]]). This is done by relating these transformations to the activity of certain bacterial or archaean species<ref name="ma"/>. | |
| − | |||
| − | |||
| − | |||
===Research in the central Baltic Sea=== | ===Research in the central Baltic Sea=== | ||
| − | |||
Studies of these transition zones (also called redoxclines) in the central Baltic Sea showed that [[chemoautotrophic]] prokaryotes dominated microbial abundance (20-40 % of total cell numbers) and production. Most of these chemoautotrophic prokaryotes have been identified as [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393209 ''Epsilonproteobacteria'']. A more detailed study of this group revealed that ''Epsilonproteobacteria'' were nearly entirely represented by the genus [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393613 ''Sulfurimonas'']. | Studies of these transition zones (also called redoxclines) in the central Baltic Sea showed that [[chemoautotrophic]] prokaryotes dominated microbial abundance (20-40 % of total cell numbers) and production. Most of these chemoautotrophic prokaryotes have been identified as [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393209 ''Epsilonproteobacteria'']. A more detailed study of this group revealed that ''Epsilonproteobacteria'' were nearly entirely represented by the genus [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393613 ''Sulfurimonas'']. | ||
This organism can be called a [[Keystone species|“key player”]] in this [[habitat]] because it mediates among other things chemoautotrophic [[denitrification]]. | This organism can be called a [[Keystone species|“key player”]] in this [[habitat]] because it mediates among other things chemoautotrophic [[denitrification]]. | ||
| − | A [[strain]] of this genus demonstrated the exceptional metabolic versatility. It could utilise different inorganic redox reactions as well as to make use of different organic substrates. This is likely to be an adaptation for survival in these [[pelagic]] redoxclines which are characterized by steep physico-chemical gradients but also by frequent disturbances due to inflow and small-scale mixing events | + | A [[strain]] of this genus demonstrated the exceptional metabolic versatility. It could utilise different inorganic redox reactions as well as to make use of different organic substrates. This is likely to be an adaptation for survival in these [[pelagic]] redoxclines which are characterized by steep physico-chemical gradients but also by frequent disturbances due to inflow and small-scale mixing events<ref name="ma"/>. |
| − | + | ||
| − | |||
| − | |||
==Microbial research in the coastal North West Mediterranean Sea== | ==Microbial research in the coastal North West Mediterranean Sea== | ||
| − | |||
===Relative microbial abundance=== | ===Relative microbial abundance=== | ||
| − | |||
[http://www.marbef.org MarBEF] researchers have investigated the relative abundance of certain microbial species in the [[Coastal_area|coastal]] North West [[Mediterranean Sea and Region, including Adriatic Sea|Mediterranean Sea]]. | [http://www.marbef.org MarBEF] researchers have investigated the relative abundance of certain microbial species in the [[Coastal_area|coastal]] North West [[Mediterranean Sea and Region, including Adriatic Sea|Mediterranean Sea]]. | ||
On an annual basis, [http://www.marinespecies.org/aphia.php?p=taxdetails&id=392750 Alphaproteobacteria] are the dominant group. The '''SAR11''' subgroup are the most abundant during spring and summer. However only a small amount (<10%) of these SAR11 bacteria take up aminoacids, glucose or ATP (an indication for activity). The '''Roseobacter''' subgroup (also from the alphaproteobacteria) is less abundant and can only be detected in winter and spring. In contrast Roseobacter cells, which make up only 5-10% of the alphaproteobacteria, are much more active, as more of them incorporate these substrates. | On an annual basis, [http://www.marinespecies.org/aphia.php?p=taxdetails&id=392750 Alphaproteobacteria] are the dominant group. The '''SAR11''' subgroup are the most abundant during spring and summer. However only a small amount (<10%) of these SAR11 bacteria take up aminoacids, glucose or ATP (an indication for activity). The '''Roseobacter''' subgroup (also from the alphaproteobacteria) is less abundant and can only be detected in winter and spring. In contrast Roseobacter cells, which make up only 5-10% of the alphaproteobacteria, are much more active, as more of them incorporate these substrates. | ||
| − | An other [[phyla|phylum]], the '''Bacteroidetes''', constitutes the second most important group and is equally abundant throughout the year. A third Phylum, [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393018 Gammaproteobacteria], showed a small peak during summer, but was only very abundant on one particular day. The [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393019 Alteromonadas] subgroup of Gammaproteobacteria constituted a population of highly active cells that were all actively taking up organic matter. However, they were quickly eliminated from the water by grazers | + | An other [[phyla|phylum]], the '''Bacteroidetes''', constitutes the second most important group and is equally abundant throughout the year. A third Phylum, [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393018 Gammaproteobacteria], showed a small peak during summer, but was only very abundant on one particular day. The [http://www.marinespecies.org/aphia.php?p=taxdetails&id=393019 Alteromonadas] subgroup of Gammaproteobacteria constituted a population of highly active cells that were all actively taking up organic matter. However, they were quickly eliminated from the water by grazers<ref name="ma"/>. |
[[Image:Bacterial abundance.jpg|thumb|centre|700px| <div> | [[Image:Bacterial abundance.jpg|thumb|centre|700px| <div> | ||
| Line 75: | Line 66: | ||
===R and K strategies=== | ===R and K strategies=== | ||
| − | |||
These results indicate that some groups of bacteria in the North West Mediterranean act as r-strategists; meaning fastgrowing opportunists. They have high rates of growth and they dominate the incorporation of the present substrates, but they are seldom abundant. These groups would be the Roseobacter and the Alteromonadacea. | These results indicate that some groups of bacteria in the North West Mediterranean act as r-strategists; meaning fastgrowing opportunists. They have high rates of growth and they dominate the incorporation of the present substrates, but they are seldom abundant. These groups would be the Roseobacter and the Alteromonadacea. | ||
<P> | <P> | ||
| − | Other groups, such as the Alphaproteobacteriaceae, SAR11 and the Bacteroidetes, follow more the alternative k-strategy, with slow growth and relative dominance | + | Other groups, such as the Alphaproteobacteriaceae, SAR11 and the Bacteroidetes, follow more the alternative k-strategy, with slow growth and relative dominance<ref name="ma"/>. |
| − | + | ||
| − | + | ||
| − | |||
==Microbial research in the deep North Atlantic== | ==Microbial research in the deep North Atlantic== | ||
| − | |||
===Deep sea microbial diversity=== | ===Deep sea microbial diversity=== | ||
| + | [http://www.marbef.org MarBEF] researchers followed the bacterial and archaeal community composition of the major [[Deep sea habitat|deep-water]] masses of the North Atlantic from 65°N to 5°S. It was found that each of the main deep-water masses is characterized by a specific bacterial community. In general, the [[Microbial_research#Determining_microbial_diversity|diversity of bacterial communities]] was about three times higher than that of the archaeal community throughout the entire water column (down to a depth of 4,500 m). It was revealed that neither bacterial nor archaeal diversity decreased with depth, although the total number of species decreased from 106, at the surface, to 104 species per ml at 4,500 m<ref name="ma"/>. | ||
| − | |||
| − | |||
| − | |||
| − | |||
===Latitudinal gradient=== | ===Latitudinal gradient=== | ||
| + | A pronounced latitudinal gradient was detected for ''Crenarchaeota''. It was previously assumed that all Marine Crenarchaeota Group I (MCGI) use ammonia as an energy source and take up carbon dioxide as a carbon source. MarBEF found that, while MCGI oxidise ammonia throughout the water column in the northern latitudes, in the deep waters around the equator only a small fraction of the MCGI utilize ammonia as an energy source. MCGI in the deep temperate and (sub)tropical waters also utilise organic matter as substrate and hence exhibit a [[heterotrophic]] life mode. | ||
| − | + | The shift from [[autotrophic]], ammonia-oxidizing northern deep water MCGI communities to heterotrophic, deep-water MCGI in equatorial regions is apparently related to the age of the deep-water masses. The deep water, which is formed in the North Atlantic, transfers large amounts of surface water ammonia into the deep ocean. This ammonia is then oxidized to nitrate as these deep waters age during the [[ocean circulation]]<ref name="ma"/>. | |
| − | + | Other ecological latitudinal patterns can be found in the article [[Ecological and latitudinal aspects]]. | |
| − | |||
| − | |||
| − | |||
| − | |||
==References== | ==References== | ||
<references/> | <references/> | ||
| − | [[Category:MarBEF | + | [[Category:MarBEF Wiki]] |
| − | [[Category: | + | [[Category:Coastal and marine ecosystems]] |
Latest revision as of 12:44, 4 April 2025
Contents
Microbes rule the world
In the ocean, microbes (organisms from 0.2 to 100µm) are very abundant. It has been calculated that they account for about half of the biomass on Earth. In the ocean, Bacteria and Archaea account for billions of tonnes of carbon (estimates range from 3 to 14 billion) while, in contrast, all people on Earth combined only account for about 0.03 billion tonnes of carbon. In a drop (one millilitre) of seawater, one can find 10 million viruses, one million bacteria and about 1,000 small protozoans and algae (called “protists”). In addition to their high abundance, microbes play a crucial role in most biogeochemical processes occurring in the marine environment: they account for almost half of global primary production and form a major part of ecosystem respiration and nutrient recycling[1].
Determining microbial diversity
Research in recent years has shown that microbes are not only very abundant and ecologically important but are also highly diverse. Although they have, in most cases, a very similar morphology, we know that they perform very different functions and that they contain very different genetic material (DNA), which is a coding for a large variety of proteins.
As scientists cannot identify most microbes from their appearance alone, they have to rely on molecular methodologies to describe their diversity. In general, these methodologies rely on the fact that microbes share a common gene that is so important that it has changed relatively little throughout the evolutionary history of life on Earth. Analysing the differences in that gene enables organisms to be classified into different taxa in a way that reflects their evolutionary history.
Initial reports of microbial richness in aquatic environments suggested that there were less than 200 different microorganisms in a typical sea water sample. But recent advances in molecular technologies have shown that the diversity is far much greater. Today it is thought that a single seawater sample may contain up to 10.000 different types of microorganisms[1].
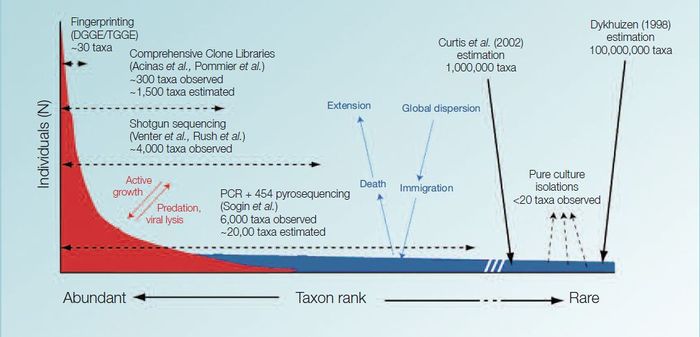
Specific discoveries
To understand plankton distribution and changes, one should know how diverse micro-plankton species are. Some fundamental questions are: ‘Can a bacterium species evolve locally despite its limited dispersal?’ and conversely, ‘Is there a bacterium species which inhabits all European waters?’
Rhodopirellula, an abundant red bacterium that lives attached to marine sediment grains was selected to investigate these questions. 70 different strains, representing several different species of the Rhodopirellula genus were isolated. Rhodopirellula baltica was restricted to the Baltic Sea, the Skagerrak and the Eastern North Sea. A second Rhodopirellula species of was present in Iceland and Scotland, representing the North-Atlantic habitat. Another Rhodopirellula species was obtained from the Adriatic Sea, but the majority of the isolates belonged to a forth Rhodopirellula species which was present in the English Channel, on the French Atlantic coast and in the Mediterranean. This showed that different species of the Rhodopirellula genus evolved within the European seas.
Molecular techniques allow the detection of the most abundant microbes. Therefore, it allows us to attempt to identify the main microbes that participate in biogeochemical cycling in different marine habitats. This therefore provides a link between biodiversity (or at least a component of it) with ecosystem functioning.
The trick is to find out “who is doing what” and “who is the most relevant” among those that perform a given biogeochemical function and what effect global change will have on that particular species.
Members of the MarBEF project have researched the identity and ecological function of the key microbial organisms in different areas[1]: the central Baltic Sea, the North West Mediterranean Sea and the deep North Atlantic.
Microbial research in the central Baltic Sea
Anoxic zones
Anoxic bottom waters are characteristic features of marginal and enclosed seas and many coastal environments (Black Sea, Baltic Sea, fjords, etc) and are increasing in extent worldwide.
In oxic-anoxic transition zones, elements are transformed from one state into another. These transformations have an impact the biogeochemical cycles and are important on an ecosystem scale. Furthermore, these environments can give insight in how ancient oceans, which were mostly anoxic throughout much of the Earth’s history, worked. These transition zones are also ideal sites to study the link between microbial community structure and biogeochemical transformations (and thus between biodiversity and ecosystem function). This is done by relating these transformations to the activity of certain bacterial or archaean species[1].
Research in the central Baltic Sea
Studies of these transition zones (also called redoxclines) in the central Baltic Sea showed that chemoautotrophic prokaryotes dominated microbial abundance (20-40 % of total cell numbers) and production. Most of these chemoautotrophic prokaryotes have been identified as Epsilonproteobacteria. A more detailed study of this group revealed that Epsilonproteobacteria were nearly entirely represented by the genus Sulfurimonas. This organism can be called a “key player” in this habitat because it mediates among other things chemoautotrophic denitrification.
A strain of this genus demonstrated the exceptional metabolic versatility. It could utilise different inorganic redox reactions as well as to make use of different organic substrates. This is likely to be an adaptation for survival in these pelagic redoxclines which are characterized by steep physico-chemical gradients but also by frequent disturbances due to inflow and small-scale mixing events[1].
Microbial research in the coastal North West Mediterranean Sea
Relative microbial abundance
MarBEF researchers have investigated the relative abundance of certain microbial species in the coastal North West Mediterranean Sea.
On an annual basis, Alphaproteobacteria are the dominant group. The SAR11 subgroup are the most abundant during spring and summer. However only a small amount (<10%) of these SAR11 bacteria take up aminoacids, glucose or ATP (an indication for activity). The Roseobacter subgroup (also from the alphaproteobacteria) is less abundant and can only be detected in winter and spring. In contrast Roseobacter cells, which make up only 5-10% of the alphaproteobacteria, are much more active, as more of them incorporate these substrates.
An other phylum, the Bacteroidetes, constitutes the second most important group and is equally abundant throughout the year. A third Phylum, Gammaproteobacteria, showed a small peak during summer, but was only very abundant on one particular day. The Alteromonadas subgroup of Gammaproteobacteria constituted a population of highly active cells that were all actively taking up organic matter. However, they were quickly eliminated from the water by grazers[1].
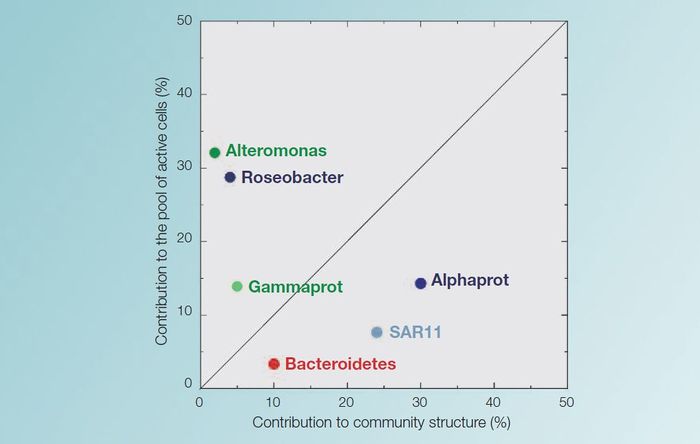
R and K strategies
These results indicate that some groups of bacteria in the North West Mediterranean act as r-strategists; meaning fastgrowing opportunists. They have high rates of growth and they dominate the incorporation of the present substrates, but they are seldom abundant. These groups would be the Roseobacter and the Alteromonadacea.
Other groups, such as the Alphaproteobacteriaceae, SAR11 and the Bacteroidetes, follow more the alternative k-strategy, with slow growth and relative dominance[1].
Microbial research in the deep North Atlantic
Deep sea microbial diversity
MarBEF researchers followed the bacterial and archaeal community composition of the major deep-water masses of the North Atlantic from 65°N to 5°S. It was found that each of the main deep-water masses is characterized by a specific bacterial community. In general, the diversity of bacterial communities was about three times higher than that of the archaeal community throughout the entire water column (down to a depth of 4,500 m). It was revealed that neither bacterial nor archaeal diversity decreased with depth, although the total number of species decreased from 106, at the surface, to 104 species per ml at 4,500 m[1].
Latitudinal gradient
A pronounced latitudinal gradient was detected for Crenarchaeota. It was previously assumed that all Marine Crenarchaeota Group I (MCGI) use ammonia as an energy source and take up carbon dioxide as a carbon source. MarBEF found that, while MCGI oxidise ammonia throughout the water column in the northern latitudes, in the deep waters around the equator only a small fraction of the MCGI utilize ammonia as an energy source. MCGI in the deep temperate and (sub)tropical waters also utilise organic matter as substrate and hence exhibit a heterotrophic life mode.
The shift from autotrophic, ammonia-oxidizing northern deep water MCGI communities to heterotrophic, deep-water MCGI in equatorial regions is apparently related to the age of the deep-water masses. The deep water, which is formed in the North Atlantic, transfers large amounts of surface water ammonia into the deep ocean. This ammonia is then oxidized to nitrate as these deep waters age during the ocean circulation[1].
Other ecological latitudinal patterns can be found in the article Ecological and latitudinal aspects.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Heip, C., Hummel, H., van Avesaath, P., Appeltans, W., Arvanitidis, C., Aspden, R., Austen, M., Boero, F., Bouma, TJ., Boxshall, G., Buchholz, F., Crowe, T., Delaney, A., Deprez, T., Emblow, C., Feral, JP., Gasol, JM., Gooday, A., Harder, J., Ianora, A., Kraberg, A., Mackenzie, B., Ojaveer, H., Paterson, D., Rumohr, H., Schiedek, D., Sokolowski, A., Somerfield, P., Sousa Pinto, I., Vincx, M., Węsławski, JM., Nash, R. (2009). Marine Biodiversity and Ecosystem Functioning. Printbase, Dublin, Ireland ISSN 2009-2539