Coral Molecular Biomarkers
Contents
An Overview of the Ecotoxicology of Coral
I. Introduction
Coral reefs worldwide are in deep trouble due to negative impacts from human activities and climate change (Wilkinson, 2004; Hughes et al., 2010; Richmond & Wolanski, 2011; Graham, 2014). In order to assess the health of coral reefs, a number of ecological monitoring programs have been developed. However, most of the current coral reef assessment and monitoring programs still largely use mortality as an indicator (e.g. loss of species and coral cover reductions), which is an “ineffective metric of stress when the desired outcome is health” (Richmond & Wolanski 2011). Traditional reef monitoring and assessment cannot determine the exact causes, as well as the relative contribution of multiple stressors (Downs et al. 2005). In order to assess the health of coral reefs more effectively (i.e. identifying the stressors, figuring out their causative effects, and detecting stress early), researchers began to use biomedical research tools to diagnose environmental health, including corals (Richmond & Wolanski 2011; Downs et al. 2005). This gave a rise to coral ecotoxicology, a relatively ‘young’ field, which studies the effects of chemicals and potential pollutants on individual organisms with the intent of applying such data to understanding their effects on populations and community structures and functions (Richmond 2004). Ecotoxicology aims to predict the effects of potential pollutants at the population, community or ecosystem level so that we can prevent or remediate possible detrimental situations effectively and efficiently. Coral ecotoxicology is designed to ‘diagnose’ coral health at a sublethal level, since stress response, such as changes in protein production and gene expression, usually occurs before physiological damage is evident (Rougée 2011; Downs et al. 2005). Coral ecotoxicological studies investigate various physiological responses to stress-inducing agents, such as protein production related to detoxification and oxidative damage, changes in growth rate, larval settlement, and bleaching (Downs et al. 2012; Ferrier-Pagès et al. 2001; Fitt et al. 2001; Negri & Heyward 2001). A number of biomarkers, or biological indicators, have been identified and used to assess the stress responses in coral. Biomarkers can be categorized into two main groups, molecular and non-molecular. The molecular markers assess changes in protein expression levels, enzyme activity levels, DNA damage, and gene expression levels. The non-molecular markers include measuring the effects on fertilization success, larval-settlement, metamorphosis, growth rates and mortality/survivorship. This article provides a comprehensive overview of the coral ecotoxicological studies conducted recently in both the laboratory and the field. First, molecular biomarkers used in coral ecotoxicological studies are summarized along with the major cellular stress-response pathways characterized in coral. Second, by shifting the focus from biomarkers to stress-inducing agents, the effects of major inducing agents on coral are summarized. Lastly, an example of a successful study utilizing the ecotoxicology technique is introduced with the summary.
II. Cellular/sub-cellular Stress Responses in Coral (Molecular Biomarkers)
A number of laboratory exposure experiments and field studies have been conducted to assess the responses to stress-inducing agents in coral using molecular and non-molecular biomarkers. The focus of this review is on molecular markers, which are often employed to detect sublethal, cellular-level responses to stress-inducing agents.
1) Protein Expression / Enzyme Activity Levels:
Changes in certain protein production or enzymatic activity have been employed as molecular markers in coral ecotoxicological studies in order to detect the sublethal stress response. Several major stress response pathways are fairly well understood in coral, and summarized below, along with the molecular biomarkers associated with these pathways.
Xenobiotic and detoxification response
Xenobiotic metabolism is characterized in a number of species of coral (Rotchell & Ostrander 2011). Coral cells contain detoxification enzymes that metabolize and expel harmful xenobiotics by altering their chemical structure, making them less toxic and more easily excreted. This generally involves a 3-phase process (Figure 1). The Phase I enzymes catalyze reactions such as hydrolysis, reduction, and oxidation of xenobiotics, and make them slightly more hydrophilic (Jokanović 2001). Cytochrome P450 monooxygenase (CYP), a large family of enzymes with broad substrate specificity, have been used to measure this Phase I activity (e.g. Downs et al. 2012; Downs et al. 2006; Rougée et al. 2006; Rotchell & Ostrander 2011). In Phase II, the conjugative reactions add the Phase I metabolites with endogenous substrates such as sulfates, acetates, glutathione, and glucuronic acid, which increase the toxicant’s water solubility and facilitate excretion (Downs, et al. 2012). Glutathione-S-transferase (GST), which conjugates glutathione to toxic metabolites, is used to measure the Phase II activity in coral (e.g. Downs et al. 2012; Downs & Downs 2006; Rougée et al. 2006). The last stage involves the transportation of the hydrophilic metabolites to lysosomes for further metabolism for containment or excretion from the cell. The biomarker for this process is multixenobiotic resistance protein (MXR). MXR is an ATP-binding cassette transporter that exports glutathione-conjugated compounds out of the cell (e.g. Downs et al. 2012; Downs et al. 2006; Rougée et al. 2005). CYP, GST and MXR are the primary biomarkers to assess the xenobiotic response in coral. Most studies use all of these biomarkers to ensure the presence of the response (Downs et al. 2012; Downs et al. 2009; Downs & Downs 2006; Downs et al. 2006; Rougée et al. 2006). Table 1 summarizes the biomarkers that are used for assessing the xenobiotic response in coral.
Examples: Significantly elevated expression of CYP (6-class) was reported in coral Pocillopora damicornis in Guam from a reef located near a golf course, compared to the reference site (Downs et al. 2012). This golf course site was known to experience elevated levels of sedimentation periodically due to run-off. Based on the elevated xenobiotic-response biomarker level, the corals at the golf course site were speculated not only experiencing sedimentation stress, but also responding to pesticide exposure from golf course run-off (Downs et al. 2012). The significantly increased xenobiotic-response biomarker , GST activity, was induced in Montastraea faveolata by exposure to high concentrations of Benzo[a]pyrene (BaP), a component of various fuels and oils (Ramos & García 2007). Significantly higher GST and MXR were also reported in P. damicornis following laboratory exposures to crude oil (Rougée et al. 2006). These results elucidate that corals possess the detoxification mechanism for petrogenic substances such as oils and fuels.
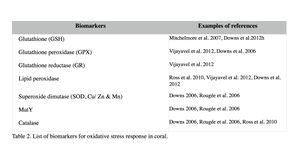
Oxidative damage and stress biomarkers
Oxidative damage/stress results from the production of reactive oxygen species such as free radicals and peroxide, and can be induced in coral by a variety of stressors, including toxic chemicals, abnormal temperatures, and UV irradiation (Rotchell & Ostrander 2011). This oxidative stress can damage lipids, proteins and DNA, and may lead to cell death if the stress continues (Downs et al. 2002). Oxidative stress response can be measured by the changes in antioxidant production or enzymatic activity level. Biomarkers used to assess oxidative stresses in coral are summarized in Table 2 (e.g. Downs et al. 2012; Vijayavel et al. 2012; Downs et al. 2011; Rotchell & Ostrander 2011; Rougée et al. 2006; Downs et al. 2002). Cells respond to the production of damaging oxygen radicals by producing antioxidant enzymes and proteins to neutralize these reactive oxygen species (Figure 2) (Rotchell & Ostrander 2011). For example, glutathione (GSH) is one of the major antioxidants in cells, and the altered GSH levels can be measured by the kinetic assay of glutathione peroxidase (GPX), glutathione reductase (GR), or the GSH content directly or indirectly (e.g. using free thiols and proteic thiol in Farina et al. 2008).
Examples: Following exposure to BaP, coral larvae of Porites astreoides showed a significant increase in GSH content (Farina et al. 2008), indicating that BaP induced oxidative stress, in addition to the xenobiotic response in coral as previously described. Excessive concentrations of iron are also known to lead to the generation of free radical species, resulting in the oxidative stress and peroxidation of membrane lipids in cells (Vijayavel et al. 2012). Oxidative stress induced by iron in larvae of P. damicornis is discussed further in Section III. Coral larvae of P. astreoides showed the oxidative stress response to a toxin produced by the dinoflagellate, Karenia brevis. K. brevis produce a series of neurotoxins and cause red tides in Florida (Ross et al. 2010). Following exposure to live cells or cellular lysates of K. brevis, the activity levels of lipid hydroperoxide and catalase, as well as concentrations of carbonyl in P. astreoides were significantly elevated, indicating oxidative stress. Although no significant reduction was observed in larval settlement and survival in this experiment (after 20-hour exposure), Ross et al. (2010) state that these coral larvae are unlikely to survive a longer exposure to the toxin of K. brevis due to the high-level of oxidative injury observed.
Protein metabolic condition
Changes in certain protein production and turnover indicate a significant shift in cellular metabolism and homeostasis (Rougée et al. 2006; Downs et al. 2006). The biomarkers for protein metabolism are listed in Table 3. Heat shock proteins (Hsp) are a chaperone that ensures the correct protein foldings, and binds to denatured proteins to prevent them from causing cell damage (Rougée et al. 2006). They are called ‘stress proteins’ since their expression is elevated by not only thermal stress but also a variety of other stressors, such as heavy metals, UV, and toxic chemicals (Santoro 2000). Elevated levels of these proteins also indicate a significant system-wide shift in the protein metabolic conditions and equilibrium (Downs et al. 2012; Downs et al. 2006; Rougée et al. 2006). Ubiquitin and ubiquitin ligase are involved in the degradation of broken or unnecessary proteins. Their up-regulation indicates that a demand for protein degradation has increased, or the process has amplified (Downs & Downs 2006; Downs et al. 2006).
Examples: Upregulation of heat shock proteins in coral was reported from a number of studies. For example, field studies conducted to assess the general health of coral reefs in the Virgin Islands (P. astreoides) and Guam (P. damicornis) report that high heat shock protein levels, along with other biomarkers, were detected from potentially polluted coral reef sites (Downs et al. 2012; Downs et al. 2011).Theses sites had higher concentrations of pesticides, herbicides and fungicides than the reference sites, indicating the overall stress levels of these reefs were higher. Gene expressions are also used to detect changes in the heat shock protein expression in coral (see II-3 for more information). For instance, increased expression levels of the Hsp 70 genes were reported after exposing Montastraea franksi to copper and an oil dispersant (Corexit 9527), and Hsp 90 gene expression was also elevated by oil dispersant exposure (Venn 2009).
Others
Other characterized stress response pathways in coral include porphyrin metabolism and homeostatic changes. These pathways are shown to be altered/disrupted by exposure to a variety of stressors. The biomarkers used to assess porphyrin metabolism (Table 4) and metabolic homeostasis (Table 5) are summarized below.
2) DNA damage
Several studies have reported DNA damage in coral from exposure to stress-inducing agents, using different techniques. The standard COMET assay (single cell gel electrophoresis) was successfully applied to quantify the increases in DNA damage after exposing Montastraea franksi to copper of different dilutions (1). Alternatively, the DNA damage can be assessed by measuring the formation of DNA adducts, which are covalent adducts formed between chemical mutagens and DNA. The DNA adducts-formation was reported from P. lobata exposed to hydrocarbon after the oil-spill incident in Yap (Downs et al. 2006). In addition, increased apurinic/apyrimidinic (AP) sites, which are DNA without a purine or a pyrimidine base due to the damage/stress, were observed as a response to exposure of P. damicornis larvae to iron chloride (Vijayavel et al. 2012). The field study sites in the Virgin Islands also showed higher AP sites in P. astreoides, where high concentrations of pollutants were detected (Downs et al. 2011). If we include environmental stressors, several more studies have reported DNA damage in coral. Physical damage to DNA was observed after exposing the cell suspensions of Stylophora pistillata to UVB irradiation (Baruch 2005). Increased AP sites were also observed by exposing S. pistillata to hyposalinity (28 ppt, 24 ppt, 20 ppt) (Downs et al. 2009). Additionally, an elevated level of MutY enzyme, the DNA glycosylase that repairs oxidative damage to DNA, may indicate the DNA damage is occurring in the cell (Downs et al. 2006).
3) Gene expression
Increasing numbers of studies have demonstrated that gene expression can evaluate the relative impact of stressors in coral. Upregulation or downregulation of gene expressions associated with stress-response pathways are reported in several studies (e.g. Edge et al. 2005). The coral genes studied as stress response to inducing agents include:
- Heat shock protein (Hsp70, Hsp90) genes - the expression was upregulated by thermal stress, Cu exposure, & oil dispersant exposure (Császár et al. 2009; Venn et al. 2009).
- Ferritin gene - the expression was upregulated by thermal stress (Császár et al. 2009).
- Superoxide dimutase (SOD-Mn) gene - the expression was upregulated by thermal stress (Császár et al., 2009).
- Zn2+-metalloprotease gene - the expression was upregulated by thermal stress (Császár et al., 2009).
- Carbonic anhydrase gene - the expression was downregulated by thermal stress, but upregulated by hypersalinity & UV (Edge et al. 2005).
- uPAR (urokinase plasminogen activator receptor) gene - the expression was upregulated by hypersalinity & UV, but the mixed response was obtained by thermal stress (Edge et al. 2005).
- P-glycoprotein (P-gp)(multi-xenobiotic resistance protein) gene - upregulated by Cu and oil dispersant exposure (Venn et al. 2009).
- Metallothionein coding gene - (this gene is known to be involved in stress response, and was successfully isolated from Acropora cervicornis, but the expression was not induced by thermal, UV or hypersalinity stress [Edge et al. 2005; Snell et al. 2003]).
Measuring stress responses using gene expressions in coral is a rapidly developing filed and will require further studies to understand the mechanism comprehensively. With an advancement of next generation sequencing (NGS) techniques, researchers now can capture the whole transcriptomic response to toxicants or other environmental stressors without specific knowledge on the mechanisms or pathways involved. However, the relationship between stress responses and gene expressions can be complex, as can be seen in the unique gene expression profiles observed in Edge et al. (2005)’s study, which showed uPAR -urokinase plasminogen activator receptor- upregulated the most at the intermediate stress level. Nevertheless, this ‘toxicogenomic’ approach has a great potential to diagnose sublethal stress in coral and advance our knowledge.
4) Other endpoints
The effects of stress-inducing agents in coral can be assessed by a variety of endpoints other than molecular biomarkers, such as fertilization success, larval settlement, metamorphosis, growth rates, and mortality/survivorship. Although non-molecular biomarkers are not the focus of this review, the major ecotoxicological responses measured by non-molecular markers are summarized in Table 7 (some of these responses will be discussed in the next section). Early life stages of many marine organisms are found to be more sensitive to pollutants than adult stages (Reichelt-Brushett & Harrison 1999). This is probably due in part to the important role that chemical cues play in substrate selection and metamorphosis of larvae in many marine organisms (Hadfield & Paul 2001). Indeed, select species of coral larvae use chemical cues to settle on a substratum where a specific coralline algal species is present (Golbuu & Richmond 2007). Therefore, it is critical to understand the effects of pollutants on gametes and larvae.