Difference between revisions of "Microbial research"
(→Specific discoveries) |
|||
| Line 38: | Line 38: | ||
Molecular techniques allow the detection of the most abundant microbes. Therefore, it allows us to attempt to identify the main microbes that participate in biogeochemical cycling in different marine habitats. This therefore provides a link between biodiversity (or at least a component of it) with ecosystem functioning. | Molecular techniques allow the detection of the most abundant microbes. Therefore, it allows us to attempt to identify the main microbes that participate in biogeochemical cycling in different marine habitats. This therefore provides a link between biodiversity (or at least a component of it) with ecosystem functioning. | ||
The trick is to find out “who is doing what” and “who is the most relevant” among those that perform a given biogeochemical function and what effect global change will have on that particular species. <ref name="ma">[http://www.marbef.org/documents/glossybook/MarBEFbooklet.pdf Heip, C., Hummel, H., van Avesaath, P., Appeltans, W., Arvanitidis, C., Aspden, R., Austen, M., Boero, F., Bouma, TJ., Boxshall, G., Buchholz, F., Crowe, T., Delaney, A., Deprez, T., Emblow, C., Feral, JP., Gasol, JM., Gooday, A., Harder, J., Ianora, A., Kraberg, A., Mackenzie, B., Ojaveer, H., Paterson, D., Rumohr, H., Schiedek, D., Sokolowski, A., Somerfield, P., Sousa Pinto, I., Vincx, M., Węsławski, JM., Nash, R. (2009). Marine Biodiversity and Ecosystem Functioning. Printbase, Dublin, Ireland ISSN 2009-2539]</ref> | The trick is to find out “who is doing what” and “who is the most relevant” among those that perform a given biogeochemical function and what effect global change will have on that particular species. <ref name="ma">[http://www.marbef.org/documents/glossybook/MarBEFbooklet.pdf Heip, C., Hummel, H., van Avesaath, P., Appeltans, W., Arvanitidis, C., Aspden, R., Austen, M., Boero, F., Bouma, TJ., Boxshall, G., Buchholz, F., Crowe, T., Delaney, A., Deprez, T., Emblow, C., Feral, JP., Gasol, JM., Gooday, A., Harder, J., Ianora, A., Kraberg, A., Mackenzie, B., Ojaveer, H., Paterson, D., Rumohr, H., Schiedek, D., Sokolowski, A., Somerfield, P., Sousa Pinto, I., Vincx, M., Węsławski, JM., Nash, R. (2009). Marine Biodiversity and Ecosystem Functioning. Printbase, Dublin, Ireland ISSN 2009-2539]</ref> | ||
| + | |||
Members of the MarBEF project [http://www.marbef.org/documents/GA/GydniaSopot2007/MarMICRO.pdf MarMicro] have researched the identity and ecological function of the key microbial organisms in different areas. These areas include: | Members of the MarBEF project [http://www.marbef.org/documents/GA/GydniaSopot2007/MarMICRO.pdf MarMicro] have researched the identity and ecological function of the key microbial organisms in different areas. These areas include: | ||
*[[Microbial research in the central Baltic Sea|The Central Baltic Sea]] | *[[Microbial research in the central Baltic Sea|The Central Baltic Sea]] | ||
*[[Microbial research in the coastal North West Mediterranean Sea|The Mediterranean Sea]] | *[[Microbial research in the coastal North West Mediterranean Sea|The Mediterranean Sea]] | ||
*[[Microbial research in the deep North Atlantic|The Deep North Atlantic]] | *[[Microbial research in the deep North Atlantic|The Deep North Atlantic]] | ||
| − | |||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 11:12, 26 August 2009
Contents
Microbes rule the world
In the ocean, microbes (organisms from 0.2 to 100µm) are very abundant. It has been calculated that they account for about half of the biomass on planet Earth. In the ocean, Bacteria and Archaea account for billions of tonnes of carbon (estimates range from 3 to 14 billion) while, in contrast, all people on Earth combined only account for about 0.03 billion tonnes of carbon. In a drop (one millilitre) of seawater, one can find 10 million viruses, one million bacteria and about 1,000 small protozoans and algae (called “protists”). In addition to their high abundance, microbes play a crucial role in most biogeochemical processes occurring in the marine environment: they account for almost half of global primary production and form a major part of ecosystem respiration and nutrient recycling.[1]
Determining microbial diversity
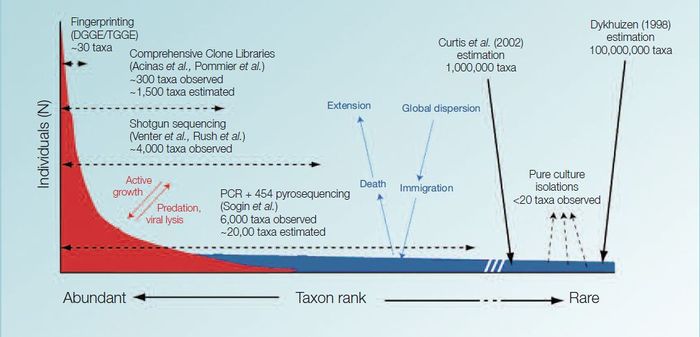
Research in recent years has shown that microbes are not only very abundant and ecologically important but are also highly diverse. Although they have, in most cases, a very similar morphology, we know that they perform very different functions and that they contain very different genetic material (DNA), which is a coding for a large variety of proteins.
As scientists cannot identify most microbes from their appearance alone, they have to rely on molecular methodologies to describe their diversity. In general, these methodologies rely on the fact that microbes share a common gene that is so important that it has changed relatively little throughout the evolutionary history of life on Earth. Analysing the differences in that gene enables organisms to be classified into different taxa in a way that reflects their evolutionary history.
Initial reports of microbial richness in aquatic environments suggested that there were less than 200 different microorganisms in a typical sea water sample. But recent advances in molecular technologies have shown that the diversity is far much greater. Today it is thought that a single seawater sample may contain up to 10,000 different types of microorganisms. [1]
Specific discoveries
To understand plankton distribution and changes, MarPLAN first needed to know how diverse micro-plankton species are. Using new techniques and partnerships, MarPLAN could answer some fundamental questions, such as: ‘Can a bacterium species evolve locally despite its limited dispersal?’ or conversely, ‘Is there a bacterium species which inhabits all European waters?’
Rhodopirellula, an abundant red bacterium that lives attached to marine sediment grains was selected to investigate these questions. 70 different strains, representing several different species of the Rhodopirellula genus were isolated. Rhodopirellula baltica was restricted to the Baltic Sea, the Skagerrak and the Eastern North Sea. A second Rhodopirellula species of was present in Iceland and Scotland, representing the North-Atlantic habitat. Another Rhodopirellula species was obtained from the Adriatic Sea, but the majority of the isolates belonged to a forth Rhodopirellula species which was present in the English Channel, on the French Atlantic coast and in the Mediterranean. This showed that there are different species of the Rhodopirellula genus present in European seas.
Molecular techniques allow the detection of the most abundant microbes. Therefore, it allows us to attempt to identify the main microbes that participate in biogeochemical cycling in different marine habitats. This therefore provides a link between biodiversity (or at least a component of it) with ecosystem functioning.
The trick is to find out “who is doing what” and “who is the most relevant” among those that perform a given biogeochemical function and what effect global change will have on that particular species. [1]
Members of the MarBEF project MarMicro have researched the identity and ecological function of the key microbial organisms in different areas. These areas include:
References
- ↑ 1.0 1.1 1.2 Heip, C., Hummel, H., van Avesaath, P., Appeltans, W., Arvanitidis, C., Aspden, R., Austen, M., Boero, F., Bouma, TJ., Boxshall, G., Buchholz, F., Crowe, T., Delaney, A., Deprez, T., Emblow, C., Feral, JP., Gasol, JM., Gooday, A., Harder, J., Ianora, A., Kraberg, A., Mackenzie, B., Ojaveer, H., Paterson, D., Rumohr, H., Schiedek, D., Sokolowski, A., Somerfield, P., Sousa Pinto, I., Vincx, M., Węsławski, JM., Nash, R. (2009). Marine Biodiversity and Ecosystem Functioning. Printbase, Dublin, Ireland ISSN 2009-2539